What happens if RNA splicing does not occur?
Not only do the introns not carry information to build a protein, they actually have to be removed in order for the mRNA to encode a protein with the right sequence. If the spliceosome fails to remove an intron, an mRNA with extra “junk” in it will be made, and a wrong protein will get produced during translation.
- Q. What is the result of RNA splicing?
- Q. What is the sequence of the spliced RNA?
- Q. How do you detect alternative splicing?
- Q. What would be the possible results of splicing error happens?
- Q. Why is RNA splicing important?
- Q. What enzyme is responsible for RNA splicing?
- Q. What are the 3 steps of RNA processing?
- Q. Why is RNA needed under splicing?
- Q. What is an example of alternative splicing?
- Q. Why alternative splicing is important?
- Q. What causes faulty splicing?
- Q. How is RNA splicing carried out in a cell?
- Q. What causes an error in the splicing of mRNA?
- Q. How are features learnt by deepcircode related to RNA splicing?
- Q. How are Back splicing and canonical linear splicings related?
Q. What is the result of RNA splicing?
Abstract. Most RNA splicing events result in intron removal from the pre-mRNA. A single RNA transcript is spliced such that the intron is removed and the two exons are seamlessly spliced together. Another kind of trans-splicing occurs when the pre-mRNA products of separate genes get together to produce a single mRNA.
Q. What is the sequence of the spliced RNA?
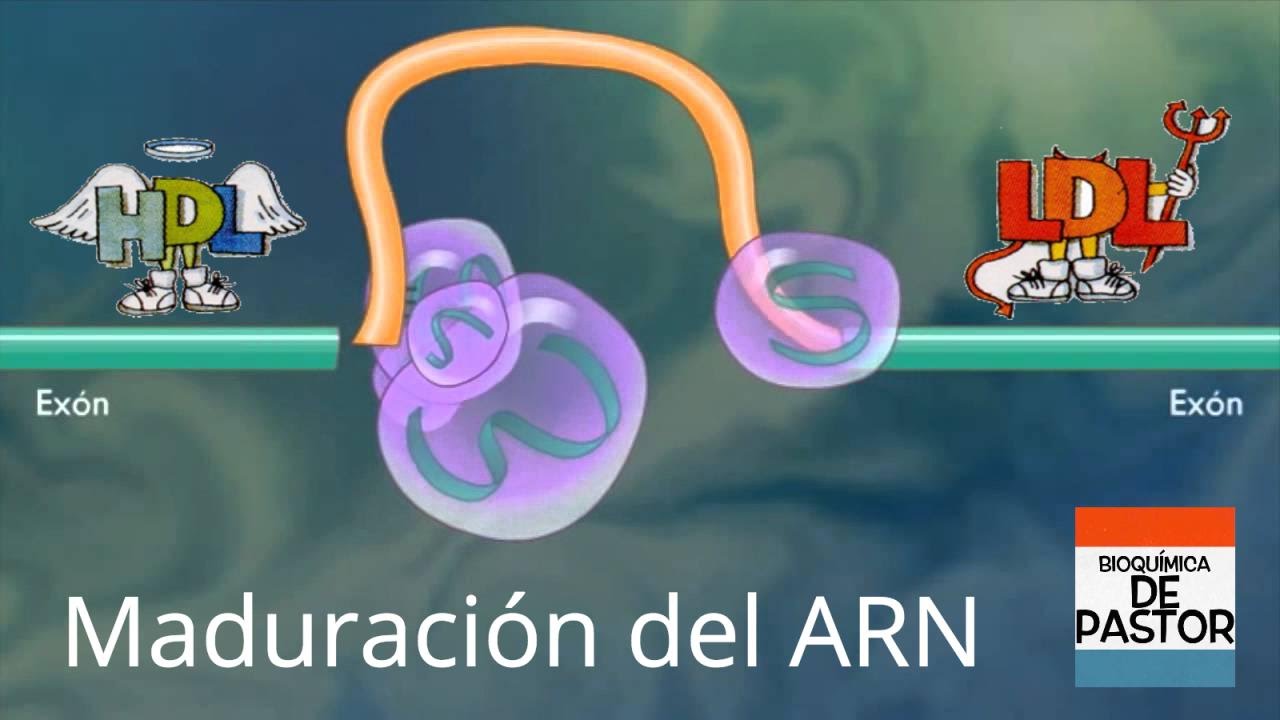
RNA splicing, in molecular biology, is a form of RNA processing in which a newly made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). During splicing, introns (non-coding regions) are removed and exons (coding regions) are joined together.
Q. How do you detect alternative splicing?
Quantification of alternative splicing to detect the abundance of differentially spliced isoforms of a gene in total RNA can be accomplished via RT-PCR using both quantitative real-time and semi-quantitative PCR methods.
Q. What would be the possible results of splicing error happens?
Any errors during the splicing process may lead to improper intron removal and thus cause alterations of the open reading frame. Therefore, the spliceosome complex has to correctly recognize and cut out the intronic sequences from the pre-mRNA molecule.
Q. Why is RNA splicing important?
Splicing makes genes more “modular,” allowing new combinations of exons to be created during evolution. Furthermore, new exons can be inserted into old introns, creating new proteins without disrupting the function of the old gene. Our knowledge of RNA splicing is quite new.
Q. What enzyme is responsible for RNA splicing?
splicing endonuclease
The RNA-splicing endonuclease is an evolutionarily conserved enzyme responsible for the excision of introns from nuclear transfer RNA (tRNA) and all archaeal RNAs.
Q. What are the 3 steps of RNA processing?
The three most important steps of pre-mRNA processing are the addition of stabilizing and signaling factors at the 5′ and 3′ ends of the molecule, and the removal of intervening sequences that do not specify the appropriate amino acids.
Q. Why is RNA needed under splicing?
Ribonucleic acid is a polymeric molecule essential in various biological roles in coding, decoding, regulation, and expression of genes. During the process of translation, the RNA undergoes splicing mechanism because it has intron and exons but introns have to be removed before the further translation takes place.
Q. What is an example of alternative splicing?
Alternative splicing is a powerful means of controlling gene expression and increasing protein diversity. The best example is the Drosophila Down syndrome cell adhesion molecule (Dscam) gene, which can generate 38,016 isoforms by the alternative splicing of 95 variable exons.
Q. Why alternative splicing is important?
Alternative splicing of RNA is a crucial process for changing the genomic instructions into functional proteins. It plays a critical role in the regulation of gene expression and protein diversity in a variety of eukaryotes. In humans, approximately 95% of multi-exon genes undergo alternative splicing.
Q. What causes faulty splicing?
Mis-regulation of splicing is a common feature of many human diseases, including several retinal diseases. Splicing defects stem from either mutation in the trans-regulatory splicing factors (Table 2) or mutations in the cis-regulatory splice site.
Q. How is RNA splicing carried out in a cell?
RNA splicing, a process exclusive to eukaryotic cells, is a post-transcriptional modification of a protein-coding messenger RNA (mRNA). This process is carried out by a complex of small nuclear RNA (snRNA) and protein, known as a spliceosome, which binds to the splice site on a pre-mRNA to fold, clip and rejoin the pre-mRNA.
Q. What causes an error in the splicing of mRNA?
Mutations that cause an error in the splicing of a messenger RNA (mRNA) can lead to diseases in humans. Various computational models have been developed to recognize the sequence pattern of the splice sites.
Q. How are features learnt by deepcircode related to RNA splicing?
Relevant features learnt by DeepCirCode are represented as sequence motifs, some of which match human known motifs involved in RNA splicing, transcription or translation. Analysis of these motifs shows that their distribution in RNA sequences can be important for back-splicing.
Q. How are Back splicing and canonical linear splicings related?
Back-splicing and canonical linear splicing can compete against each other, and some splicing factors such as Muscleblind may interact with the flanking introns to promote exon circularization ( Ashwal-Fluss et al., 2014 ).
Adquiere nuestro libro aquí:https://www.bioquimicadepastor.com/libroYa no habrá guías individuales de cada tema, ahora si quieres obtener el capítulo de Amin…
No Comments